Project-2: SCEPTRE
Last updated: 2022-08-25
Checks: 7 0
Knit directory: rotation2/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20220607) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 4d8f9d5. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Unstaged changes:
Modified: .RData
Modified: .Rhistory
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/project_2.Rmd) and HTML
(docs/project_2.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 268c781 | chenh19 | 2022-08-25 | Build site. |
| html | d5675d8 | chenh19 | 2022-08-25 | Build site. |
| html | 6f9b2d0 | chenh19 | 2022-08-25 | Build site. |
| html | eb7f6e2 | chenh19 | 2022-08-25 | Build site. |
| Rmd | df6fd70 | chenh19 | 2022-08-25 | wflow_publish("./analysis/*.Rmd") |
| html | 06d8f5b | chenh19 | 2022-08-25 | Build site. |
| Rmd | 8195470 | chenh19 | 2022-08-25 | wflow_publish("./analysis/*.Rmd") |
| html | 481c95e | chenh19 | 2022-08-25 | Build site. |
| Rmd | 0617d83 | chenh19 | 2022-08-25 | wflow_publish("./analysis/*.Rmd") |
| Rmd | 4c40591 | chenh19 | 2022-08-25 | update |
| html | 4c40591 | chenh19 | 2022-08-25 | update |
| html | 210ce87 | chenh19 | 2022-08-25 | Build site. |
| Rmd | 949442a | chenh19 | 2022-08-25 | wflow_publish("./analysis/*.Rmd") |
| html | 7126807 | chenh19 | 2022-08-25 | Build site. |
| html | e576eee | chenh19 | 2022-08-25 | Build site. |
| html | 3b152ba | chenh19 | 2022-08-25 | Build site. |
| Rmd | 140545b | chenh19 | 2022-08-25 | wflow_publish("./analysis/*.Rmd") |
| html | abd8577 | chenh19 | 2022-08-25 | Build site. |
| Rmd | 5e9089f | chenh19 | 2022-08-25 | wflow_publish("./analysis/*.Rmd") |
| html | fb0804c | chenh19 | 2022-08-25 | Build site. |
| Rmd | ddc528d | chenh19 | 2022-08-25 | wflow_publish("./analysis/*.Rmd") |
| html | 46c0eee | chenh19 | 2022-08-25 | Build site. |
| Rmd | 0f18210 | chenh19 | 2022-08-25 | wflow_publish("./analysis/*.Rmd") |
| html | 4bcd287 | Hang Chen | 2022-08-11 | Build site. |
| html | 316e143 | Hang Chen | 2022-08-11 | Build site. |
| Rmd | 52c15a2 | Hang Chen | 2022-08-11 | wflow_publish("./analysis/*.Rmd") |
| html | 2ecff7a | Hang Chen | 2022-08-11 | Build site. |
| Rmd | ba2bb58 | Hang Chen | 2022-08-11 | wflow_publish("./analysis/*.Rmd") |
| html | 1a7329a | Hang Chen | 2022-08-11 | Build site. |
| html | d98b8ed | Hang Chen | 2022-08-10 | Build site. |
| html | afbc7f7 | chenh19 | 2022-08-09 | Build site. |
| Rmd | 0193b6d | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | f3ddb60 | chenh19 | 2022-08-09 | Build site. |
| Rmd | a977d05 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | aeeaff4 | chenh19 | 2022-08-09 | Build site. |
| Rmd | 8a2aa07 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | faa4371 | chenh19 | 2022-08-09 | Build site. |
| Rmd | 03c1f34 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | 40d7285 | chenh19 | 2022-08-09 | Build site. |
| Rmd | e21456b | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | 51ff922 | chenh19 | 2022-08-09 | Build site. |
| Rmd | 9c70e05 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | 60bb670 | chenh19 | 2022-08-09 | Build site. |
| Rmd | 74886cf | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | 5035de6 | chenh19 | 2022-08-09 | Build site. |
| Rmd | 877bc81 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | c710b98 | chenh19 | 2022-08-09 | Build site. |
| Rmd | ae2d7c0 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | 9c8a125 | chenh19 | 2022-08-09 | Build site. |
| Rmd | babe576 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | 759a524 | chenh19 | 2022-08-09 | Build site. |
| Rmd | 117a594 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | 0137730 | chenh19 | 2022-08-09 | Build site. |
| Rmd | ce281e3 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | 306629d | chenh19 | 2022-08-09 | Build site. |
| Rmd | 7169d04 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | a2f14fc | chenh19 | 2022-08-09 | Build site. |
| Rmd | 142e870 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | afe9e5e | chenh19 | 2022-08-09 | Build site. |
| Rmd | dd9c995 | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | 7f87abe | chenh19 | 2022-08-09 | Build site. |
| Rmd | 8f127ae | chenh19 | 2022-08-09 | wflow_publish("./analysis/*.Rmd") |
| html | f7a30e6 | chenh19 | 2022-08-08 | Build site. |
| Rmd | 75aba85 | chenh19 | 2022-08-08 | wflow_publish("./analysis/*.Rmd") |
| html | ae6dc22 | chenh19 | 2022-08-08 | Build site. |
| Rmd | af06843 | chenh19 | 2022-08-08 | wflow_publish("./analysis/*.Rmd") |
| html | 31345e3 | chenh19 | 2022-08-08 | Build site. |
| html | ab573d7 | chenh19 | 2022-08-08 | Build site. |
| Rmd | 7f24442 | chenh19 | 2022-08-08 | wflow_publish("./analysis/*.Rmd") |
| html | 82d0b8a | chenh19 | 2022-08-08 | Build site. |
| Rmd | aff1e3c | chenh19 | 2022-08-08 | wflow_publish("./analysis/*.Rmd") |
| html | 63022aa | chenh19 | 2022-08-08 | Build site. |
| Rmd | 8808ac1 | chenh19 | 2022-08-08 | wflow_publish("./analysis/*.Rmd") |
| html | b4ec414 | chenh19 | 2022-08-08 | Build site. |
| html | fb4fa31 | chenh19 | 2022-08-08 | Build site. |
| Rmd | 7c18d6a | chenh19 | 2022-08-08 | wflow_publish("./analysis/*.Rmd") |
| html | 03df33f | chenh19 | 2022-08-08 | Build site. |
| Rmd | e784a3f | chenh19 | 2022-08-08 | wflow_publish("./analysis/*.Rmd") |
| html | ec5763a | chenh19 | 2022-08-08 | Build site. |
| html | 870ec95 | chenh19 | 2022-08-08 | Build site. |
| html | 4df0f61 | chenh19 | 2022-08-08 | Build site. |
| html | 8bad269 | chenh19 | 2022-08-08 | Build site. |
| html | 6e10041 | chenh19 | 2022-08-08 | Build site. |
| html | b045b81 | chenh19 | 2022-08-08 | Build site. |
| html | e22ab6b | chenh19 | 2022-08-08 | Build site. |
| html | 87b3f9d | chenh19 | 2022-08-08 | Build site. |
| html | 78b6bd6 | chenh19 | 2022-08-08 | Build site. |
| html | 60fabb8 | Hang Chen | 2022-08-08 | Build site. |
| html | cee42b8 | Hang Chen | 2022-08-05 | Build site. |
| html | 6927e45 | Hang Chen | 2022-08-04 | Build site. |
| html | 551a34f | Hang Chen | 2022-08-04 | Build site. |
| html | 80908a7 | Hang Chen | 2022-08-04 | Build site. |
| html | 2623d6b | Hang Chen | 2022-08-04 | Build site. |
| html | e9d9966 | Hang Chen | 2022-08-04 | Build site. |
| html | 57d96a8 | Hang Chen | 2022-08-04 | update |
| Rmd | 05b3310 | Hang Chen | 2022-08-04 | update |
| html | 05b3310 | Hang Chen | 2022-08-04 | update |
| html | 37d15c9 | chenh19 | 2022-07-19 | Build site. |
| Rmd | 3576593 | chenh19 | 2022-07-19 | wflow_publish("./analysis/*.Rmd") |
| html | 8f6816e | chenh19 | 2022-07-19 | Build site. |
| Rmd | 383342f | chenh19 | 2022-07-19 | wflow_publish("./analysis/*.Rmd") |
| html | 4a94b94 | chenh19 | 2022-07-19 | Build site. |
| Rmd | a18fc1f | chenh19 | 2022-07-19 | wflow_publish("./analysis/*.Rmd") |
| html | 870115f | chenh19 | 2022-07-19 | Build site. |
| html | 9241fe6 | chenh19 | 2022-07-08 | Build site. |
| html | 0a7633b | chenh19 | 2022-07-07 | Build site. |
| html | 63535ad | chenh19 | 2022-07-07 | Build site. |
| html | feb5923 | chenh19 | 2022-07-07 | Build site. |
| html | c849244 | chenh19 | 2022-07-07 | Build site. |
| html | 7ab5d71 | chenh19 | 2022-07-07 | Build site. |
| html | 625e7ca | chenh19 | 2022-07-07 | Build site. |
| html | 0e1d92d | chenh19 | 2022-07-07 | Build site. |
| html | 24d6bcf | chenh19 | 2022-07-07 | Build site. |
| html | 3a14766 | chenh19 | 2022-07-07 | Build site. |
| html | ca77db2 | chenh19 | 2022-07-07 | Build site. |
| html | e0cb7a2 | chenh19 | 2022-06-28 | Build site. |
| html | bf05c98 | chenh19 | 2022-06-28 | Build site. |
| html | 7cfb685 | chenh19 | 2022-06-28 | Build site. |
| html | 43dad9c | chenh19 | 2022-06-28 | Build site. |
| html | 7aa3172 | chenh19 | 2022-06-28 | Build site. |
| html | 2254608 | chenh19 | 2022-06-28 | Build site. |
| html | 968a4b6 | chenh19 | 2022-06-23 | Build site. |
| html | 229f924 | chenh19 | 2022-06-23 | Build site. |
| html | 9750134 | chenh19 | 2022-06-23 | Build site. |
| html | 4b38bf6 | chenh19 | 2022-06-23 | Build site. |
| html | 8a7980d | chenh19 | 2022-06-23 | Build site. |
| html | 3e1478e | chenh19 | 2022-06-22 | Build site. |
| html | 5258612 | chenh19 | 2022-06-22 | Build site. |
| html | 2382868 | chenh19 | 2022-06-22 | Build site. |
| html | 1144127 | chenh19 | 2022-06-22 | Build site. |
| html | da2c0fe | chenh19 | 2022-06-21 | Build site. |
| html | 8aa5960 | chenh19 | 2022-06-21 | Build site. |
| html | 6783fa3 | chenh19 | 2022-06-21 | Build site. |
| html | d9be701 | chenh19 | 2022-06-21 | Build site. |
| html | 82e6e50 | chenh19 | 2022-06-21 | Build site. |
| html | d376ad0 | chenh19 | 2022-06-21 | Build site. |
| html | 325a212 | chenh19 | 2022-06-21 | Build site. |
| html | e14c55c | chenh19 | 2022-06-21 | Build site. |
| html | f971bbd | chenh19 | 2022-06-21 | Build site. |
| html | dca882f | chenh19 | 2022-06-21 | Build site. |
| html | 3a80eaf | chenh19 | 2022-06-21 | Build site. |
| html | 33829a5 | chenh19 | 2022-06-21 | Build site. |
| html | 5b446cf | chenh19 | 2022-06-21 | Build site. |
| html | 28a06ee | chenh19 | 2022-06-21 | Build site. |
| html | d5b4ff0 | chenh19 | 2022-06-21 | Build site. |
| html | 16400a7 | chenh19 | 2022-06-21 | Build site. |
| html | a324166 | chenh19 | 2022-06-21 | Build site. |
| html | 27a4d51 | chenh19 | 2022-06-21 | Build site. |
| html | f765024 | chenh19 | 2022-06-21 | Build site. |
| html | bc55dbb | chenh19 | 2022-06-21 | Build site. |
| html | b3b7ed6 | chenh19 | 2022-06-21 | Build site. |
| html | 405d57e | chenh19 | 2022-06-21 | Build site. |
| html | c10a1a8 | chenh19 | 2022-06-21 | Build site. |
| html | 2443e38 | chenh19 | 2022-06-21 | Build site. |
| Rmd | 8e0c8ad | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | 5d192d2 | chenh19 | 2022-06-21 | Build site. |
| Rmd | 356550c | chenh19 | 2022-06-21 | wflow_publish("./analysis/*.Rmd") |
| html | 21e9501 | chenh19 | 2022-06-21 | Build site. |
| html | b002776 | chenh19 | 2022-06-20 | Build site. |
| html | 6211c60 | chenh19 | 2022-06-15 | Build site. |
| html | 0da18e2 | chenh19 | 2022-06-15 | Build site. |
| html | 0aff555 | chenh19 | 2022-06-15 | Build site. |
| html | a4e1e73 | chenh19 | 2022-06-15 | Build site. |
| html | f0e98f9 | chenh19 | 2022-06-14 | Build site. |
| html | eafa16b | chenh19 | 2022-06-14 | Build site. |
| html | dfd60ce | chenh19 | 2022-06-14 | Build site. |
| Rmd | 49f1922 | chenh19 | 2022-06-14 | wflow_publish("./analysis/*.Rmd") |
| html | fd7271e | chenh19 | 2022-06-14 | Build site. |
| html | 1b4d12e | chenh19 | 2022-06-14 | Build site. |
| html | a6c402d | chenh19 | 2022-06-14 | Build site. |
| html | cedad99 | chenh19 | 2022-06-14 | Build site. |
| html | e3b9788 | chenh19 | 2022-06-14 | Build site. |
| html | aed5eed | chenh19 | 2022-06-14 | Build site. |
| html | 45bb6ed | chenh19 | 2022-06-14 | Build site. |
| html | c23765a | chenh19 | 2022-06-14 | Build site. |
| Rmd | 1277c93 | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | b5fb71c | chenh19 | 2022-06-14 | Build site. |
| html | 152325b | chenh19 | 2022-06-14 | Build site. |
| html | 543ef4c | chenh19 | 2022-06-14 | Build site. |
| html | 9b6cb27 | chenh19 | 2022-06-14 | Build site. |
| html | 8674c8a | chenh19 | 2022-06-14 | Build site. |
| html | ada4068 | chenh19 | 2022-06-14 | Build site. |
| html | 0d08121 | chenh19 | 2022-06-14 | Build site. |
| html | dd2046d | chenh19 | 2022-06-14 | Build site. |
| html | 2c4cab1 | chenh19 | 2022-06-14 | Build site. |
| html | d46eaab | chenh19 | 2022-06-14 | Build site. |
| html | 02a0c26 | chenh19 | 2022-06-14 | Build site. |
| html | abad46e | chenh19 | 2022-06-14 | Build site. |
| html | 741027b | chenh19 | 2022-06-14 | Build site. |
| html | bb15812 | chenh19 | 2022-06-14 | Build site. |
| html | 93a27ae | chenh19 | 2022-06-14 | Build site. |
| html | 27121a9 | chenh19 | 2022-06-14 | Build site. |
| Rmd | fce1ffd | chenh19 | 2022-06-14 | wflow_publish("analysis/*.Rmd") |
| html | 44517c1 | chenh19 | 2022-06-13 | Build site. |
| html | da08f11 | chenh19 | 2022-06-13 | Build site. |
| html | 572f6ba | chenh19 | 2022-06-13 | Build site. |
| html | b8870d3 | chenh19 | 2022-06-13 | Build site. |
| html | 719925e | chenh19 | 2022-06-13 | Build site. |
| html | e7541fa | chenh19 | 2022-06-13 | Build site. |
| html | 9d9615d | chenh19 | 2022-06-13 | Build site. |
| html | bbd8978 | chenh19 | 2022-06-13 | Build site. |
| html | e5a5b52 | chenh19 | 2022-06-13 | Build site. |
| Rmd | c43ae1f | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | 4d8bd72 | chenh19 | 2022-06-13 | Build site. |
| html | 3373521 | chenh19 | 2022-06-13 | Build site. |
| html | af21ea8 | chenh19 | 2022-06-13 | Build site. |
| Rmd | 6e56d75 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | f653f7b | chenh19 | 2022-06-13 | Build site. |
| html | d69c892 | chenh19 | 2022-06-13 | Build site. |
| html | 34d877d | chenh19 | 2022-06-13 | Build site. |
| html | e72400b | chenh19 | 2022-06-13 | Build site. |
| html | c411223 | chenh19 | 2022-06-13 | Build site. |
| html | 1daccd2 | chenh19 | 2022-06-13 | Build site. |
| Rmd | 63f46d2 | chenh19 | 2022-06-13 | wflow_publish("analysis/*.Rmd") |
| html | 26adb45 | chenh19 | 2022-06-13 | Build site. |
| html | a6022a8 | chenh19 | 2022-06-13 | Build site. |
| html | 9abc4b8 | chenh19 | 2022-06-13 | Build site. |
| html | f18d385 | chenh19 | 2022-06-13 | Build site. |
| html | e991f56 | chenh19 | 2022-06-13 | Build site. |
| html | 3c9b1d9 | chenh19 | 2022-06-13 | Build site. |
| html | 34e0d02 | chenh19 | 2022-06-13 | Build site. |
| html | 9be31af | chenh19 | 2022-06-13 | Build site. |
| html | 0f41de8 | chenh19 | 2022-06-13 | Build site. |
| html | 31ad035 | chenh19 | 2022-06-13 | Build site. |
| html | bdf3b44 | chenh19 | 2022-06-13 | Build site. |
| html | 8d0890c | chenh19 | 2022-06-13 | Build site. |
| Rmd | 26f455b | chenh19 | 2022-06-13 | update |
| html | 26f455b | chenh19 | 2022-06-13 | update |
1. Understand SCEPTRE
a. Read the paper
Some key ideas:
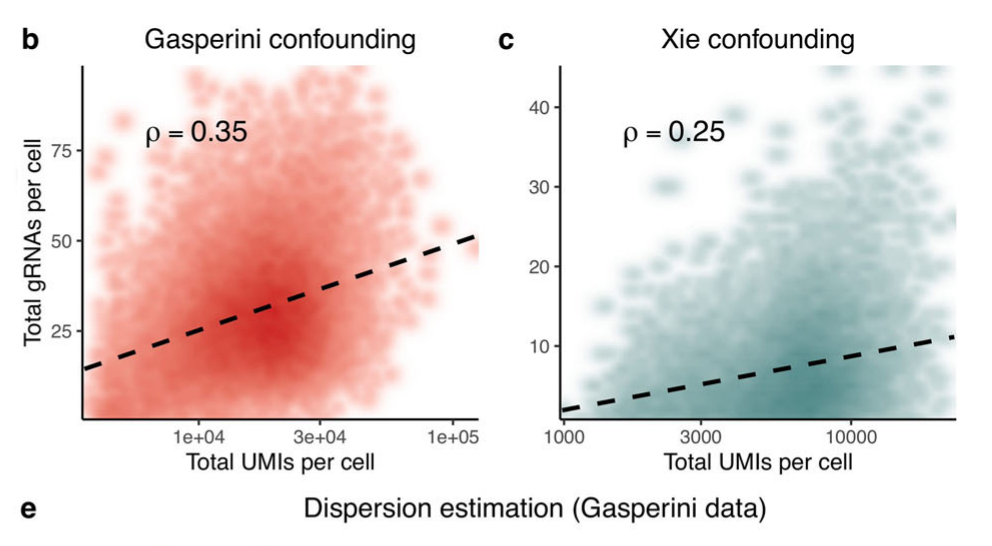
- A key confounder for perturb-seq is the read depth:
total gRNA per cellseems to show a non-linear increase astotal UMI per cellincreases.
- To overcome this, Barry et al. adopted two main strategies:
- Include depth as a covariate when doing negative binomial (NB) regression for gRNA and expression.
- Generate an empirical null distribution by randomization test.
Pipeline:
- Negative binomial regression to get z-values for each gRNA-Expression pairs.
- Logistic regression to get gRNA detection probabilities for each gRNA in each cell.
- Conditional randomization test to get the empirical null distribution (when doing randomization, use the gRNA detection probabilities as the conditions).
- Fit a skew-t distribution to the null distribution and then calculate p-values from z-values.
b. Check the package
c. Install the package
Code: sceptre.sh
d. Tutorial
e. QQ plot function
Code: qqunif.plot.R
Ref: Code
Sample: Generating QQ Plots in R
2. Manual regression
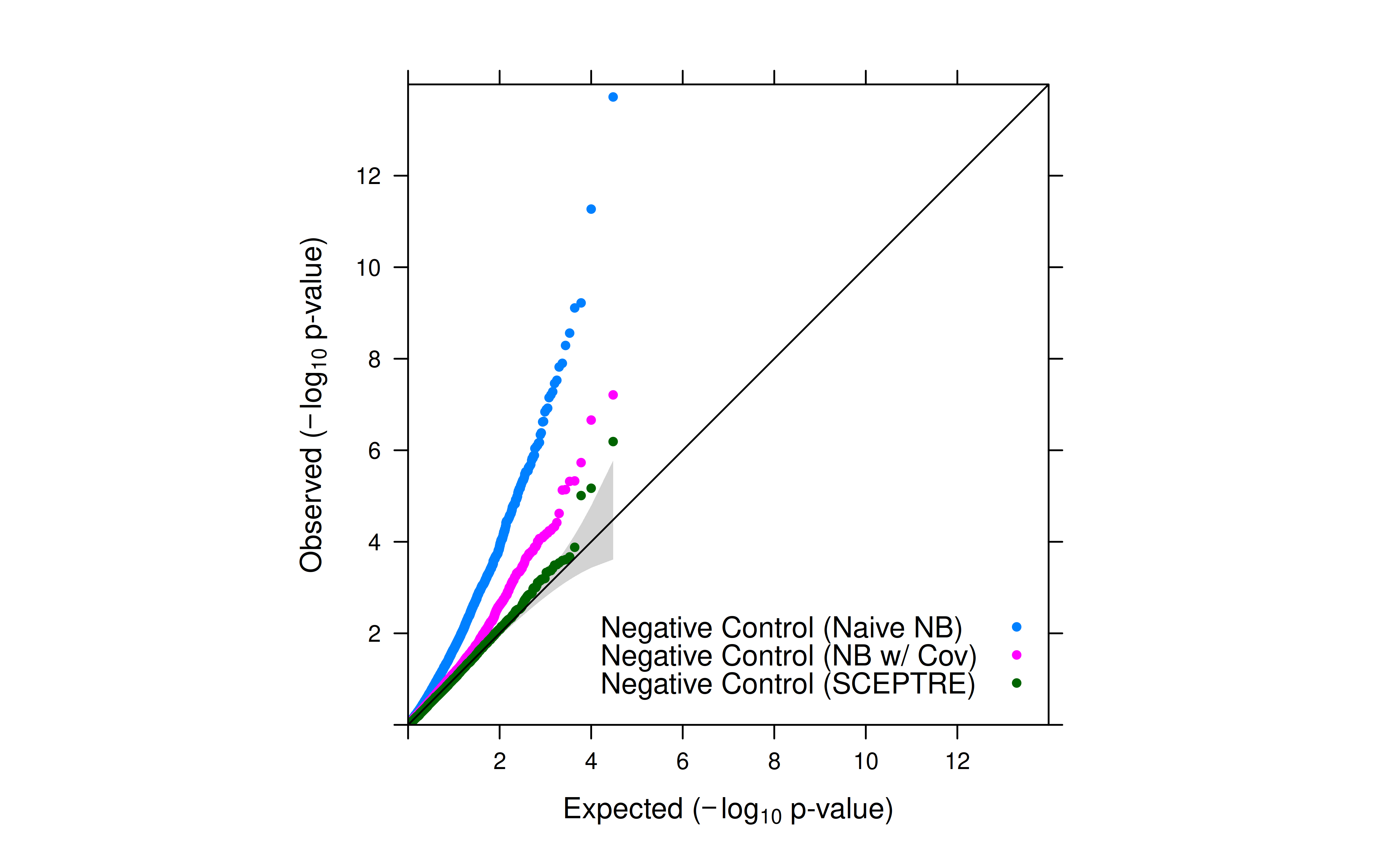
Aim: To better understand SCEPTRE, I’ll first perform naive negative binomial regression with and without total UMI as a covariate. This is mainly to check whether the negative controls have inflated p-values.
gRNA-gene pairs:
b. NB regression w/ total UMI cov
- Total UMI counts for each cell in gene_matrix was used as a covariate.
Code: NB_with_covariate.R
c. Compare
Data: p_value.zip
QQ plot
Code: negative_control_qq.R
Observation:
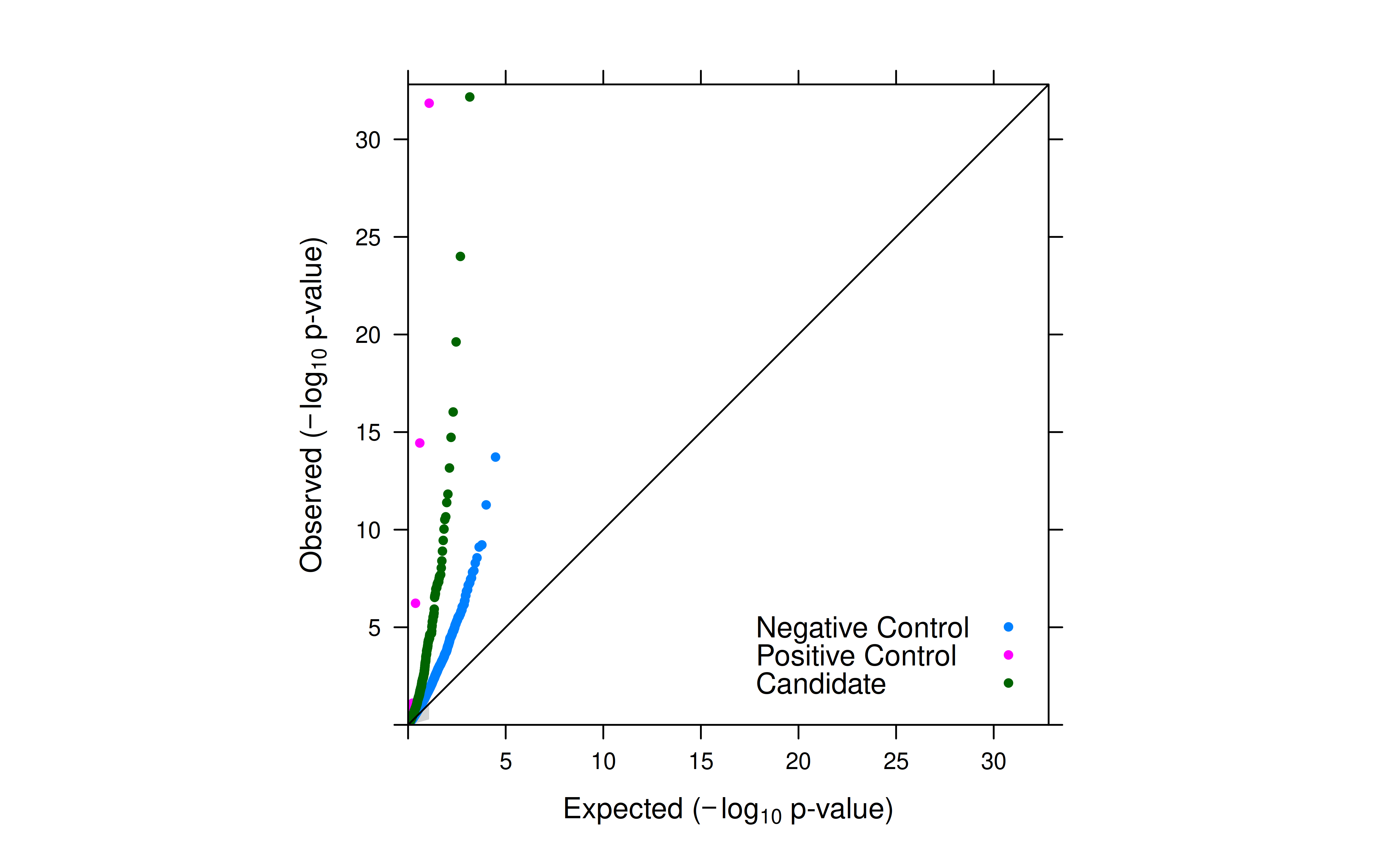
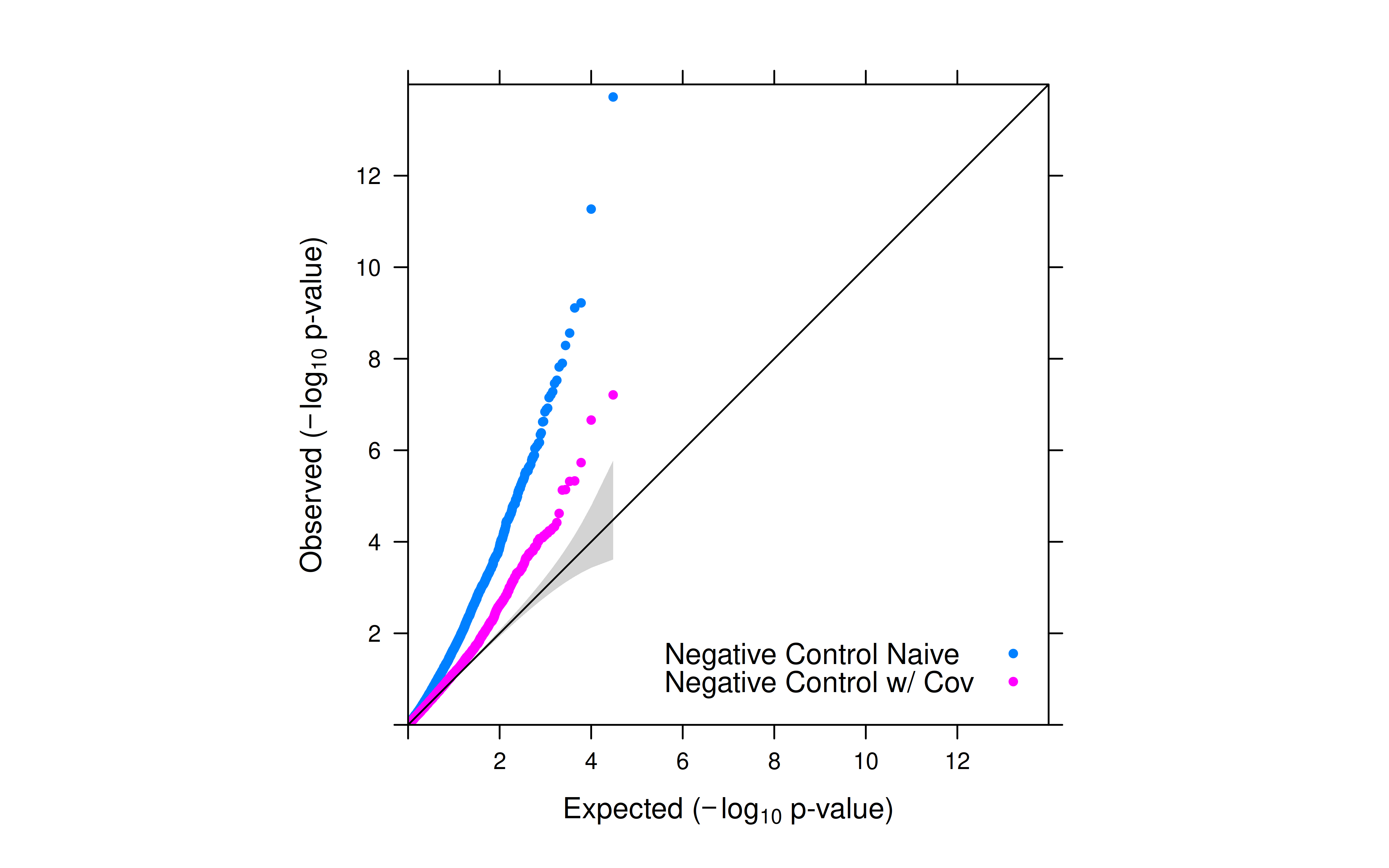
- After introducing total UMI as a covariate, the inflation for negative controls was significantly reduced.
- However, inflation is still observed. SCEPTRE’s empirical null distribution for p-value calculation is expected to further reduce the inflation.
Smallest p-values
Code: head_pvalue.R
Observation:
- For negative controls, the p-values increases after introducing the covariate in NB regression (less significant).
- For positive controls and candidates, the p_values decreases after introducing the covariate in NB regression (more significant).
[1] "The smallest p-values in [negative_control_pvalues.csv] :"
Gene gRNA p_value
1 TIMP1 NTC-2 1.893138e-14
2 DAAM1 NTC-10 5.380489e-12
3 ANAPC11 NTC-12 5.970695e-10
4 OST4 NTC-2 7.803250e-10
5 NDUFS5 NTC-12 2.727219e-09
6 MYDGF NTC-12 5.158344e-09
[1] "The smallest p-values in [negative_control_pvalues_with_covariate.csv] :"
Gene gRNA p_value
1 DAAM1 NTC-10 6.169608e-08
2 HSPA8 NTC-3 2.177909e-07
3 TPT1 NTC-3 1.876122e-06
4 TAF10 NTC-10 4.655719e-06
5 ZFAND2A NTC-3 4.809740e-06
6 VKORC1 NTC-3 7.248893e-06[1] "The smallest p-values in [positive_control_pvalues.csv] :"
Gene gRNA p_value
1 CD46 CD46-2 1.404819e-32
2 HSPA8 HSPA8-1 3.658421e-15
3 CD52 CD52-1 5.918623e-07
4 HSPA8 HSPA8-2 7.758467e-02
5 NMU NMU-1 1.077502e-01
6 PPIA PPIA-1 2.090641e-01
[1] "The smallest p-values in [positive_control_pvalues_with_covariate.csv] :"
Gene gRNA p_value
1 HSPA8 HSPA8-1 7.940576e-42
2 CD46 CD46-2 4.646680e-36
3 HSPA8 HSPA8-2 3.540640e-19
4 CD52 CD52-1 2.594172e-08
5 NMU NMU-1 5.694585e-02
6 PPIA PPIA-1 9.576647e-01[1] "The smallest p-values in [candidate_pvalues.csv] :"
Gene gRNA p_value
1 CR1L SNP-20-2 6.805935e-33
2 PTPRC SNP-14-2 1.010292e-24
3 CTU2 SNP-35-1 2.420409e-20
4 ANK1 SNP-61-1 9.305671e-17
5 KDELR2 SNP-85-1 1.871454e-15
6 PAXX SNP-29-2 6.924731e-14
[1] "The smallest p-values in [candidate_pvalues_with_covariate.csv] :"
Gene gRNA p_value
1 CR1L SNP-20-2 3.905370e-43
2 ANK1 SNP-61-1 2.579855e-31
3 PTPRC SNP-14-2 1.613814e-30
4 GLRX5 SNP-58-2 2.276511e-17
5 PDLIM1 SNP-77-2 1.042396e-12
6 NUDT4 SNP-62-2 1.123020e-123. SCEPTRE regression
Code
Code: sceptre_package_modified.zip
Code: sceptre_morris.R
Code: qqplot_sceptre_all.R
Code: top_hits.R
Note:
- I modified the SCEPTRE package source code a little bit and built the package locally.
- SCEPTRE has very strict requirements for input. For example, every
gRNA_groupmust contain 2 or more gRNAs. - I didn’t have the
covariate_matrixfrom the authors, so I just usedrnorm()to generate a matrix that I think will minimally affect the results.
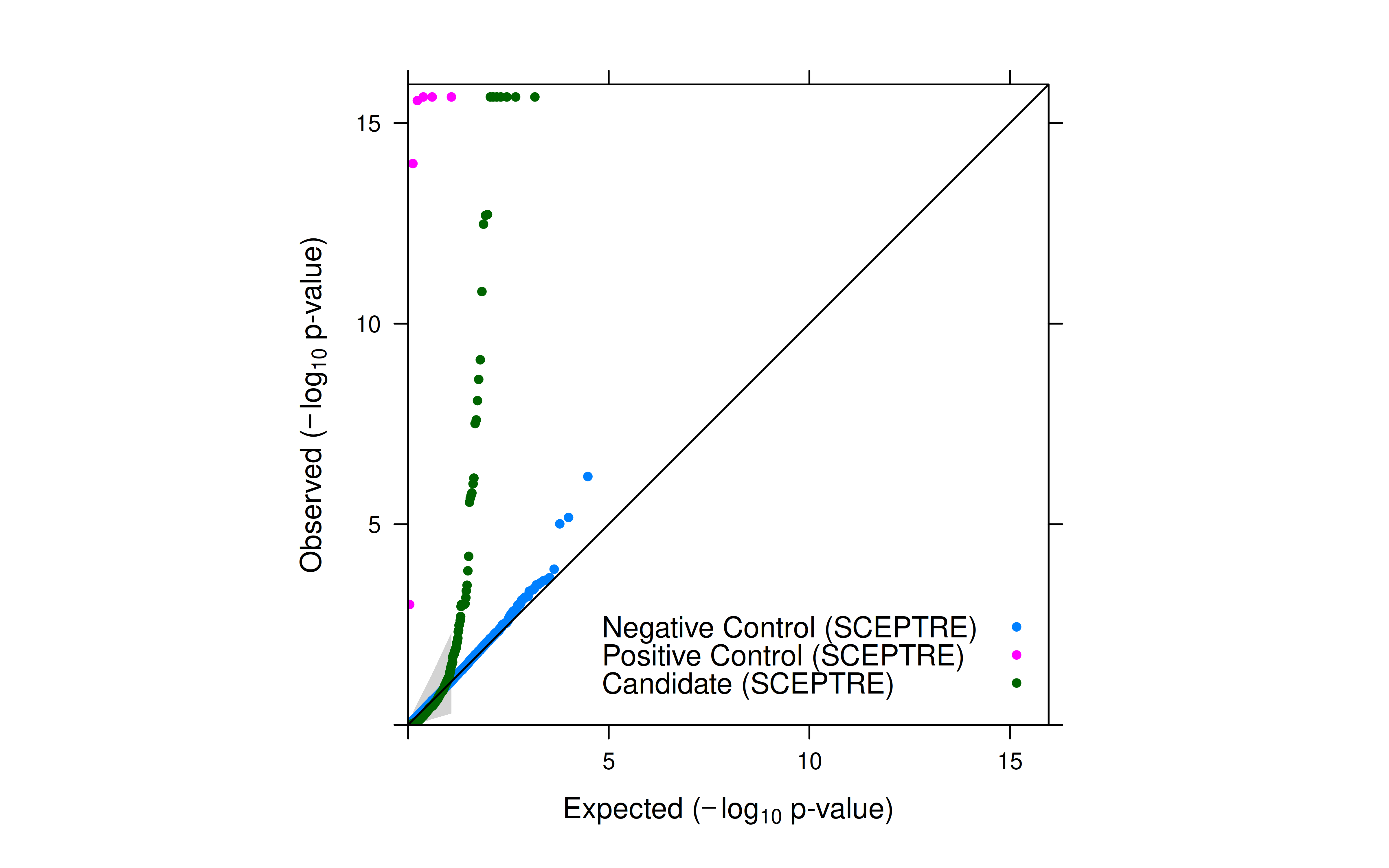
Results
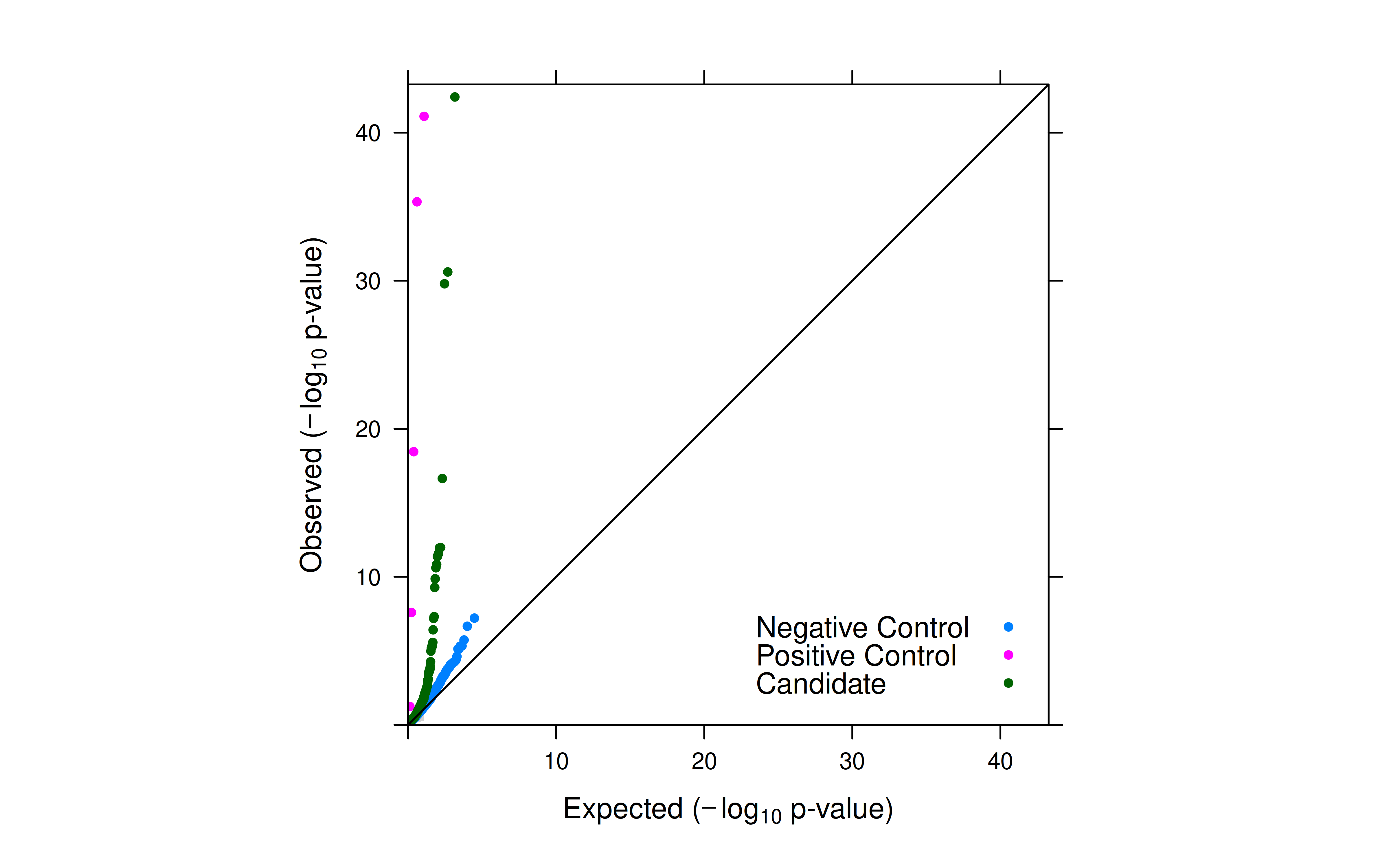
Observation
- SCEPTRE reduced inflation to a minimal level.
- Top hits from my run were all in Morris’ results (Table S3E).
Future direction
- According to Nikita, there is indeed no
covariate_matrixfrom the author. - Also according to Nikita, after PCA, there was no clear batch
separation. We cannot infer batch from the data itself, and we may just
drop the
batchcolumn incovariate_matrix. - Although no
covariate_matrixprovided by the author, I may generate it by myself. I can calculate the UMI counts, percent-mito, etc (there are 5 covariates in total might be used according to Nikita). - After running with the new
covariate_matrix, I would expect to see even less inflation in the negative controls and more significant hits.
sessionInfo()R version 4.2.1 (2022-06-23)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.1 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.10.0
LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.10.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] workflowr_1.7.0
loaded via a namespace (and not attached):
[1] Rcpp_1.0.9 compiler_4.2.1 pillar_1.8.0 bslib_0.3.1
[5] later_1.3.0 git2r_0.30.1 jquerylib_0.1.4 tools_4.2.1
[9] getPass_0.2-2 digest_0.6.29 jsonlite_1.8.0 evaluate_0.15
[13] tibble_3.1.7 lifecycle_1.0.1 pkgconfig_2.0.3 rlang_1.0.2
[17] cli_3.3.0 rstudioapi_0.13 yaml_2.3.5 xfun_0.31
[21] fastmap_1.1.0 httr_1.4.3 stringr_1.4.0 knitr_1.39
[25] sass_0.4.1 fs_1.5.2 vctrs_0.4.1 rprojroot_2.0.3
[29] glue_1.6.2 R6_2.5.1 processx_3.6.1 fansi_1.0.3
[33] rmarkdown_2.14 callr_3.7.0 magrittr_2.0.3 whisker_0.4
[37] ps_1.7.1 promises_1.2.0.1 htmltools_0.5.2 ellipsis_0.3.2
[41] httpuv_1.6.5 utf8_1.2.2 stringi_1.7.6